In January 2024, the team led by Professor Liu Zhipeng, State Key Laboratory of Herbage Improvement and Grassland Agro-ecosystems and the College of Pastoral Agricultural Science and Technology, published a paper entitled “MODMS: A Multi-omics Database for Facilitating Biological Studies in Alfalfa (Medicago sativaL.)” in Horticulture Research online.
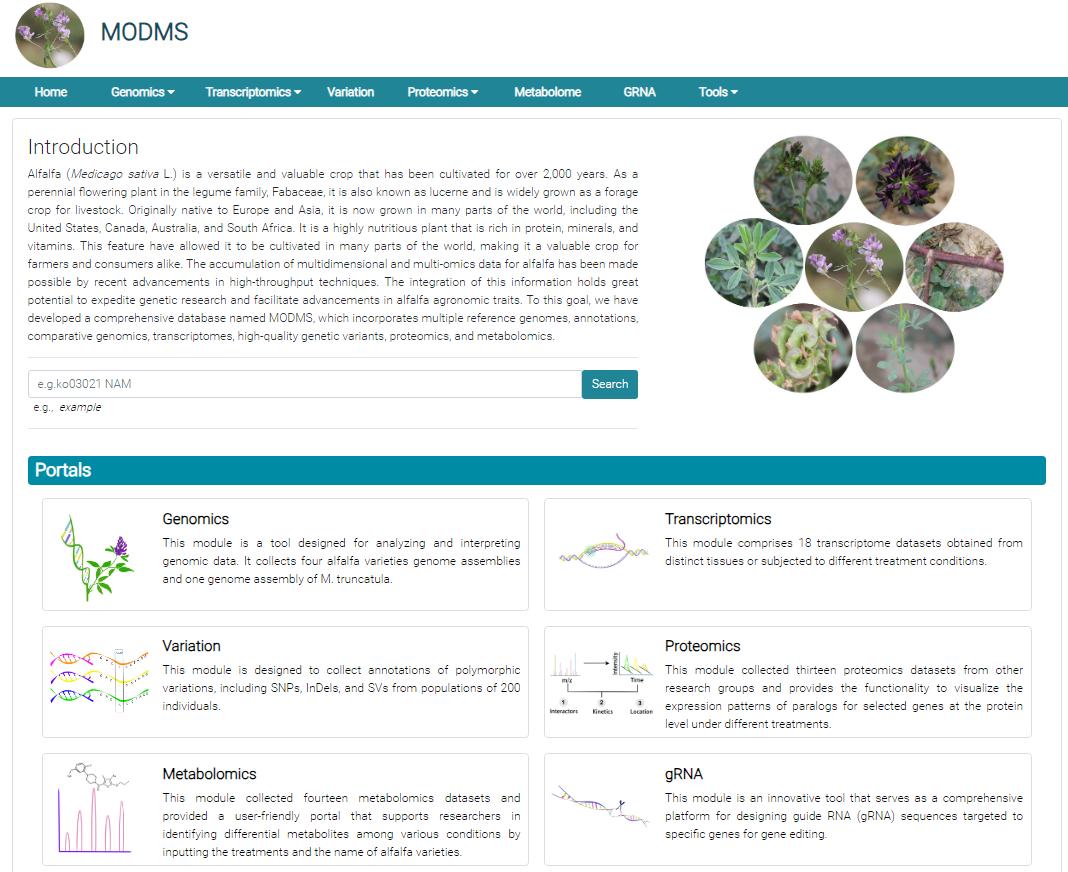
The research team developed a comprehensive multi-omics database of Medicago sativa (MODMS), which is free, user-friendly, multifunctional and incorporates previously published reference genomes, annotations, and high-quality genomic variants to query single nucleotide polymorphisms (SNPs), insertions, and deletions (InDels), as well as structural variations (SVs) of individual genes, transcriptomes, proteomics, and metabolomics for online analysis, assisting users in constructing gene regulatory networks from multiple perspectives. Additionally, MODMS integrates gene-editing tools to design single gRNA sequences, promoting the application of editing techniques in functional studies of alfalfa. By far, MODMS is the most comprehensive multi-omics database of alfalfa and the largest forage legume multi-omics database in the world.
Young researcher Fang Longfa is the first author of the paper, and Professor Liu Zhipeng is the corresponding author. The research was financially supported by the National Key Research and Development Program (2022YFF1003200) and the National Natural Science Foundation of China (Grant Nos. 32201463 and 32201444).
The construction and use of this online analysis platform is expected to provide convenience for researchers of alfalfa molecular breeding domestically and internationally.
PAPER LINK:https://doi.org/10.1093/hr/uhad245
MODMS:https://modms.lzu.edu.cn/




